Code Club S02E01: An introduction to R (Part 1)
Learning objectives
- Learn what Code Club is all about
- Get some basic familiarity with R and RStudio
- Understand a bit about R objects and how to use them
To do beforehand
Before the Code Club Zoom session, please follow the Code Club Computer Setup instructions.
In brief, you should have R and RStudio installed on your computer or you should be set up to run RStudio Server at the Ohio Supercomputer Center (OSC). (As a bonus, you can try to install and load the tidyverse package as the setup page suggests, but no sweat if you can’t get that to work yet.)
In case you run into issues, contact Jelmer or for last-minute troubleshooting, you can join the Zoom call 15-30 minutes early.
(Re)Introducing Code Club
OSU Code Club is a regularly occurring online gathering to improve coding skills, now in its second year.
This Code Club was inspired by a paper in PLoS Computational Biology (“Ten simple rules to increase computational skills among biologists with Code Clubs”), and here are some of the underlying ideas:
-
Coding is best learned by doing, so Code Club is interactive and hands-on.
-
Ongoing exposure and practice also helps when learning.
-
We aim to keep it informal and maybe even fun.
-
We have a core group of 5 organizers that do most of the presenting, but we also encourage participants to present, and will have a couple of participant-led sessions at the end of this semester (see the schedule).
Organizers
- Jelmer Poelstra (Molecular and Cellular Imaging Center (MCIC), Wooster Campus)
- Jessica Cooperstone (Dept. of Horticulture and Crop Science & Dept. of Food Science and Technology)
- Michael Broe (Dept. of Evolution, Ecology and Organismal Biology)
- Mike Sovic (Center for Applied Plant Sciences)
- Stephen Opiyo (MCIC, Columbus Campus)
Session structure
Each session consists of an instructional part where you can code along or listen, some exercises in breakout rooms with 3-4 people, and exercise recaps with the entire group.
Zoom guidelines
-
We very much welcome questions at any time, so please either unmute yourself and speak, or post in the chat whenever you have a question!
-
Having your camera on helps, especially in breakout rooms. We will record the whole-group part of each session, so we understand if some of you prefer to have their cameras off during that part. (But note that we will only share the recordings with other Code Clubbers.)
-
You can use the icons under the “Participants” menu in Zoom when we ask for a “show of hands” or if you are having problems.
-
In breakout rooms:
- Briefly introduce yourselves.
- Have someone share their screen, preferably one of the least experienced people.
- Be friendly and patient, keep everyone aboard.
- The Zoom
Ask for helpbutton will alert us, and one of the organizers will come into the breakout room. (Raise handis not seen by us outside of the room.)
Otherwise
-
We need your feedback! Always feel free to email one of the organizers, and for suggestions for a topic to cover in a future Code Club, you can fill out this form!
-
I will quickly show the Code Club menu and BioDASH website during the session.
-
Zoom polling question: are you working at OSC or locally?
1 – Why R?
R is a programming language that is most well-known for being excellent for statistical analysis and data visualization.
While the learning curve is steeper than for most programs with graphical user interfaces (GUIs), it pays off to invest in learning R:
-
R gives you greater flexibility to do anything you want.
-
Writing computer instructions as code, like you have to do in R, is more reproducible than clicking around in a GUI. It also makes it much easier to redo analyses with slight modifications!
-
R is highly interdisciplinary and can be used with many different kinds of data. To just name two examples, R has a very strong ecosystem for bioinformatics analysis (“Bioconductor” project), and can be used to create maps and perform GIS analyses.
-
R is more than a platform to perform analysis and create figures. R Markdown combines R with a simple text markup language to produce analysis reports that integrate code, results, and text, and to create slide decks, data dashboards, websites, and even books! In the third session of Code Club, Michael Broe will talk more about R Markdown.
-
While not as versatile outside of data-focused topics as a language like Python, R can be used as a general programming language, for instance to automate tasks such as large-scale file renaming.
Finally, R:
-
Is open-source and freely available for all platforms (Windows, Mac, Linux).
-
Has a large and welcoming user community.
2 – Exploring RStudio
R simply provides a “console” (command-line interface) where you can type your commands.
However, because you want to save your commands in scripts and see the graphics that you produce, it is more effective to work in an environment that provides all of this side-by-side. We will use RStudio, an excellent graphical environment (“Integrated Development Environment”, IDE) for R.
I will now demonstrate how to start an RStudio Server session from the Ohio Supercomputer Center’s website following the steps from our Code Club Computer Setup page. If you have RStudio installed on your own computer, start it now, and otherwise, follow along with me to run RStudio in your browser.
Once you have a running instance of RStudio, create a new R script by clicking File > New File > R Script.
Now, you should see all 4 “panes” that the RStudio window is divided into:
- Top-left: The Editor for your scripts and other documents (hidden when no file is open).
- Bottom-left: The R Console to interactively run your code (+ a tab with a Terminal).
- Top-right: Your Environment with R objects you have created (+ several other tabs).
- Bottom-left: Tabs for Files, Plots, Help, and others.

So, in RStudio, we have a single interface to write code in text files or directly in the console, visualize plots, navigate the files found on our computer, and inspect the data we are working with.
RStudio has a lot of useful features and during the next few sessions of Code Club, we will introduce some tips and tricks for working with it.
Breakout rooms I (~5 min.)
Introduce yourselves!
We’ll return to the same breakout room configuration later in this session to do a few exercises, so please take a moment to introduce yourself to your breakout roommates. Make sure to also mention:
-
Your level of experience with R and other coding languages.
-
What you are aiming to use or are already using R for.
Check that everyone has RStudio working
-
Take a moment to explore the RStudio interface.
-
If you run into issues, click the
Ask for helpbutton in Zoom and one of us will come by.
3 – Interacting with R
R as a calculator
The lower-left RStudio pane, i.e. the R console, is where you can interact with R directly.
The > sign is the R “prompt”. It indicates that R is ready for you to type something.
Let’s start by performing a division:
203 / 2.54
#> [1] 79.92126R does the calculation and prints the result in the console as well. Afterwards, you get your > prompt back. (The [1] may look a bit weird when there is only one output element; this is how you can keep count of output elements when there are many.)
With the expected set of symbols, you can use R as a general calculator:
203 * 2.54 # Multiplication
#> [1] 515.62
203 + 2.54 # Addition
#> [1] 205.54Note that pressing the up arrow key will put your previous command back on the prompt, and you can press the up arrow again to go further back (as well as the down arrow to go in the other direction).
Experimenting a bit…
What if we add spaces around our values?
203 - 2.54
#> [1] 200.46This works: as it turns out, R simply ignores any extra spaces.
Similarly, we could omit the single spaces around the mathematical operators that we used earlier (though we will keep using them for clarity):
203/2.54
#> [1] 79.92126How about:
203 /Now the prompt turned into a + instead of the usual >.
What is going on here? (Click for the answer)
R is waiting for you to finish the command, since you typed an incomplete command: something has to follow the division sign /.
While it was obvious here that our command was incomplete, you will often type incomplete commands without realizing you did so. Just remember that when you see the + prompt, something has to be missing in your command: most commonly, you’ll have forgotten a closing parenthesis ) or you accidentally opened up an unwanted opening parenthesis (.
If you want to abort completing the incomplete command, you can press Esc.
And if we just type a number:
203
#> [1] 203R will print the number back to us! It turns out that the default, implicit action that R will perform on anything you type is to print it back to us (under the hood, it is calling a function called print()).
Instead of a number, what if we try to have R print some text (a character string) back to us?
Fantastic
#> Error in eval(expr, envir, enclos): object 'Fantastic' not foundCode Club
#> Error: <text>:1:6: unexpected symbol
#> 1: Code Club
#> ^What seems to be going wrong here? (Click for the answer)
Whenever you type a character string, R expects to find an object with that name (we will get to what exactly objects are in a little bit!). When no object exists with that name, R will throw an error. We will learn some of the basics of objects in section 5 of today’s session.
We can get R to print character strings back to us, and work with strings in other ways, as long as we quote them:
"Fantastic"
#> [1] "Fantastic"4 – Working with a script
Need for scripts
We can go along like this, typing commands directly into the R console. But to keep better track of what we’re doing, it’s a good idea to write code in plain text files, i.e. to write “scripts”.
-
You should have already created a script above (otherwise, click
File>New File>R Script). -
Click
File>Save Asto save the script; give it a descriptive name likeintro-to-R.R.
(You may want to put the script in a new subfolder for this Code Club session.)
Interacting with the R console from your script
We recommend that you generally type your commands into a script and execute the commands from there, instead of typing directly into the console.
We want to make sure to save our division command, so start by typing the following into the R script in the top-left pane:
203 / 2.54With the cursor still on this line, press Ctrl + Enter. The command will be copied to the R console and executed, and then the cursor will move to the next line.
Note that it doesn’t matter where on the line your cursor is: Ctrl + Enter will execute the entire line unless you have selected only part of it.
(And when you have selected multiple lines of code, Ctrl + Enter will execute them all.)
Commenting
You can use # signs to annotate (comment) your code. Anything to the right of a # is ignored by R, meaning it won’t be executed. You can use # both at the start of a line or anywhere in a line following code.
Comments are a great way to describe what your code does within the code itself, so comment liberally in your R scripts! This is useful not only for others that you may share your code with, but perhaps especially for yourself when you look back at your code a day, a month, or a year later.
# Divide by 2.54 to get the wingspan in inches:
203 / 2.54 # Original measurement was in cm5 – R Objects
Assigning stuff to R objects
We can assign any value, character, or set of values or characters to an object with the assignment operator, <-. (This is a smaller-than sign < followed by a dash -.)1
For example:
wingspan_cm <- 203
conversion <- 2.54Type that into your script, and use Ctrl + Enter to send it to the console.
The objects you create get added to your “workspace” or “environment.” RStudio shows this in the Environment tab in the topright panel – check to see if wingspan_cm and conversion are indeed there.
After you’ve assigned a number to an object, you can use it in other calculations:
wingspan_inch <- wingspan_cm / conversion
wingspan_inch
#> [1] 79.92126More generally speaking, the object name that you provide is substituted with its contents by R, so the object name is just a reference to the underlying value.
Our objects so far contained just a single number and we may have also called them variables. Object is the more general name that encompasses R items of any size or complexity. As we see will see next week, R distinguishes between different types of objects.
Object names
Objects can be given any name such as x, current_temperature, or subject_id.
Some pointers on object names:
-
Because R is case sensitive,
wingspan_inchis different fromWingspan_inch. -
An object name cannot contain a space, so for readability, separate words using:
_– e.g.wingspan_inch(this is called “snake case”, which we will tend to use in Code Club instructional materials).– e.g.wingspan.inch- capitalization – e.g.
wingspanInchorWingspanInch(“camel case”)
-
Object names can contain but cannot start with a number (
2xis not valid, butx2is)2. -
Make object names descriptive yet not too long.
You will make things easier for yourself by naming objects in a consistent way, for instance by always sticking to your favorite case like “snake case."3
Objects, your workspace, and closing R
When you close R, it will probably ask you whether you want to save your workspace (“Save workspace image to ~/.RData”). When you do so, then the next time you start R, you can reload everything the way it was, such as your previously created objects.
While this may seem convenient, we recommend that you don’t do this.
Can you think of a reason why saving and reloading your workspace may not be a good idea? (Click for the answer)
The main reason why this is generally not considered good practice relates to the idea that you should be able to reproduce your workspace (and more broadly speaking, your analysis) from the code in your script.
Remember that you can modify your workspace either by entering commands in the console directly, or by running them from a script – or even from multiple different scripts. Also, in practice, you often run lines in the script out of order, or write lines in the script that you don’t execute.
Therefore, if you “carry around” the same workspace across multiple different sessions, you run a greater risk of not having a reproducible set of steps in your script.
To make RStudio stop asking you about saving your workspace, click Tools > Global Options > General and set the options as follows:
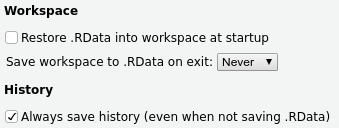
Taking these ideas a step further, it can be a good idea to occasionally restart R so you can check whether the code in your script is correct and complete, that you are not relying on code that is not in the script, and so on. To do so, you don’t need to close and reopen RStudio itself: under Session in the top menu bar, you can click Restart R (and you should also see the keyboard shortcut for it in the menu bar, which is Ctrl + Shift + F10 for Windows/Linux, and Cmd + Shift + F10 for Mac).
Breakout rooms II (5-10 min.)
Note that in both of these exercises, the answers are not contained in what we just discussed. I would like you to think about your intuition for R’s behavior, and then see if R indeed works that way or not.
Exercise 1: Object “linkage”
What do you think the value of y will be after executing the following lines in R? 100 or 160, and why?
x <- 50 # x is now 50
y <- x * 2 # y is now 100
x <- 80 # x is now 80, but what is y?Solution (click here)
Objects don’t get linked to each other, so if you change one object, it won’t affect the values of other objects that were defined earlier.
Therefore, y will continue to be 100.
Exercise 2: Errors… (Bonus)
In section 3, you might have noticed that we got a different error when typing one versus multiple unquoted words. Here are those examples again:
Fantastic
#> Error in eval(expr, envir, enclos): object 'Fantastic' not foundCode Club
#> Error: <text>:1:6: unexpected symbol
#> 1: Code Club
#> ^Reproduce these errors for yourself: in Rstudio’s editor pane, type these or equivalent error-generating examples in a script saved with a .R extension, and send them to the console.
Why is the error in the second case different, and what does it mean?
Hints (click here)
-
Can you see how RStudio can “notice” errors already in the editor – but only for the second of these two examples? The editor checks for syntax (“R grammar”) errors but not whether objects already exist.
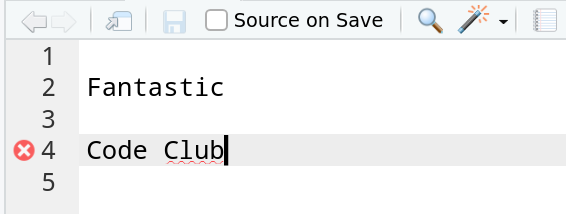
If you hover over the red cross in the margin, you can see what RStudio is upset about.
-
What if we put a
+or another operator between the two words?Code + Club #> Error in eval(expr, envir, enclos): object 'Code' not found
Solution (click here)
When typing a single unquoted word which is not an existing object, R will look for an object and then complain that it can’t find that object.
When typing multiple unquoted words with a space between them, regardless of whether those are existing objects, R will notice a syntax (“R grammar”) error before it even gets around to checking objects.
The problem is that you are referring to two objects sequentially and without any mathematical operator in between them, or some other syntax to “join” them. In R, that’s not valid syntax. (You may think it would perhaps simply try to print both objects, but this is not the case.)
A general lesson here is that you should always pay attention to the details of the error messages that you get. While the language may seem terse and odd at first, it usually holds important clues as to what is going wrong exactly.
In closing
Where to go from here
For a list of recommended resources for learning R, see our R Resources and Tips page page.
Attribution
This was modified after material from The Carpentries, especially from this Data Carpentry workshop and this “R for Ecology” workshop.
-
In RStudio, typing Alt + - will write
<-in a single keystroke. You can also use=as assignment, but that symbol can have other meanings, so we recommend sticking with the<-combination. ↩︎ -
There are some names that cannot be used because they are the names of fundamental keywords in R (e.g.,
if,else,for, see here for a complete list). In general, it’s best not to use other function names even if it’s “allowed” (e.g.,c,T,mean,data,df,weights). If in doubt, check the Help to see if the name is already in use. ↩︎ -
It is also recommended to use nouns for variable names, and verbs for function names. For more, two popular R style guides are Hadley Wickham’s and Google’s. ↩︎