Code Club:
R – Getting Started and Some Tips
New to R?
If you are completely new to R, we recommend watching at least the first couple of videos from Mike Sovic’s Youtube playlist of short videos on R, and ideally all of them, prior to attending Code Club. Here is the first video:
In case you want to do more self-study (note that this is not required/needed), here are some additional resources:
- A useful and fun written tutorial is R for cats.
- For a more systematic and lengthy introduction to R, see A Tutorial Introduction to R (this gets fairly advanced after section 9).
- Excellent comprehensive introductions are the R Basics and Visualization classes by Rafael Irizarry that can be freely accessed; you do have to create an account.
- OSU TDAI videos:
Also, don’t hesitate to reach out to the Code Club organizers if you have any questions!
Miscellaneous R tips
Useful settings
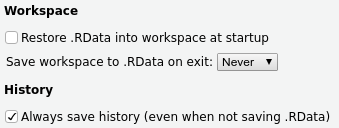
By default, R will try to save your “environment” (e.g., your loaded data, variables, etc) when you exit, and then reload everything the way it was upon restarting R. However, this is bad! You should always be able to reproduce your environment given a set of commands saved in an R script or R Markdown document, whereas saving and reloading your environment encourages you to be sloppy about this.
To disable this in RStudio, go to Tools > Global Options > General and set the options
as follows:
To start R in the same way from the command line:
R --no-save --no-restore-data
Installing R packages
CRAN packages
To install an R package that is available at CRAN, the default R package repository,
from within R (e.g. in the R console in RStudio), use the install.packages() function.
The install.packages() function will handle dependencies within R – i.e., it will install other R packages
that your package depends on. Occasionally, when the install function needs to compile
a package from source, errors arise that relate to missing system dependencies (i.e. software outside of R).
On Mac and Linux, these system dependencies are best installed outside of R,
such as with homebrew on Mac or apt on Ubuntu.
The errror message you got when trying to install an R package should tell you which system dependencies are needed.
On Windows, you can use the installr package to install such dependencies or other software from within R – for example:
install.packages("installr") # Install the installr package first
installlr::install.RStudio() # Install RStudio
installr::install.python() # Install Python
System setup to installing packages “from source”
Sometimes you need to install a package from source, that is, you need to compile the package rather than simply installing a pre-existing binary. (On Linux, where installing from source is often needed, this should work without additional steps.) On Windows and Mac, installing from source is generally only needed when you install a package from outside of CRAN (such as from Github, see below), but you will need to make sure you have the following non-R software:
On Windows, you will need Rtools (Rtools installation instructions).
On a Mac, you will need Xcode (which can be installed from the Mac App store).
You can test whether or not you are able to install packages from source using the devtools package:
install.packages("devtools") # Install the devtools package
devtools::has_devel() # Check whether you can install packages from source
For a bit more info, see this page.
Installing packages from Github
To install a package from Github, use either the devtools or the remotes package – for example:
install.packages("devtools") # Install the devtools package
devtools::install_github("kbroman/broman") # Install from a repository using "<username>/<repo-name>"
This will install the package from source, so you will need to make sure you are able to do so by following the instructions in the section right above this one.
Installing packages from Bioconductor
If you’re doing bioinformatic analyses in R, you will probably run into packages that are not on CRAN but on Bioconductor. To install a package from Bioconductor, use the BiocManager package – for example:
install.packages("BiocManager") # Install the BiocManager package
BiocManager::install("edgeR") # Install the edgeR package from Bioconductor
Updating R
Consider updating R if you have an older version of R installed.
Specifically, in the first session of Code Club, we’ve seen problems when installing
the tidyverse with R versions below R 3.6.
You can check which version of R you have by looking at the first lines of output when running the following command inside R:
sessionInfo()
To update:
-
Windows: You can update R from within R. The
updateR()function will also take care of updating your packages:install.packages("installr") installr::updateR() -
Mac: Download and install the latest
.pkgfile as if you were installing it for the first time. -
Linux: In Ubuntu, if you installed R with
aptorapt-get, you can useapt-get upgradein a terminal. Otherwise, download and install the latest version after removing the old one. Rtask has some instructions for upgrading to R 4.0 in Ubuntu (along with upgrading to Ubuntu 20.04).
Re-installing your packages after updating (Mac and Linux)
While the installr::updateR() function for Windows users takes care of reinstalling
your packages along with updating R,
Mac and Linux users will have to manually re-install their packages.
Some people prefer to re-install these packages on the fly, which can end up being a way
to get rid of packages you no longer use.
But if you want immediately reinstall all your packages, run this before you upgrade:
my_packages <- installed.packages()
saveRDS(my_packages, "my_packages.rds")
Then, after you’ve installed the latest R version:
my_packages <- readRDS("CurrentPackages.rds")
install.packages(my_packages[1, ])
This will only work for packages available on CRAN. Of course, you can check your list for Github-only and Bioconductor packages and then install those with their respective commands (see below). Yes, this can be a bit of a hassle!